The Comeback of Grapefruit: Now Safe for Medication Users

Goldenberg, L. (livnat@volcani.agri.gov.il), Carmeli-Weissberg, M., Shaya, F., Plesser, E., Kelly, G., Yaniv, Y., Arad, T., Ophir, R., Sherman, A., Carmi, N., and Eyal, Institute of Plant Sciences, ARO Volcani Center.
Background: Grapefruit-Drug Interactions
A variety of health benefits have been attributed to grapefruit; however, consumption of grapefruit is discouraged for the users of a wide range of prescription drugs, such as statins, blood thinners,anti-anxiety medications, antihistamines, and more, due to a phenomenon termed “grapefruit-drug interactions”.Notably, the use of medications affected by grapefruit consumption is very widespread and documented to stand at 35% of the general population in the USA for statins alone in 2019. Grapefruit-drug interactions are often manifested as an overdose of the drug due to delayed drug breakdown caused by furanocoumarin compounds present in large quantities in grapefruit. In other cases, consumption of grapefruit and the resulting activity of furanocoumarins can cause lower absorption of drugs, leading to a drug underdose. These compounds are present in all citrus fruits, except mandarins and oranges; however, the key genes involved in their biosynthesis have not been discovered yet.
In the current study, we identified and characterized a key gene directing furanocoumarin (FC) biosynthesis in citrus fruit/leaves, and discovered that this gene is disrupted in mandarins and oranges, which are FC-free. Our work provides the basis for developing citrus varieties which willbe safe for consumption by a large and growing population ofhumans dependent upon the use of prescription drugs.
Study Design
In order to identify candidate genes encodingkey enzymes involved in furanocoumarin biosynthesis, we created two citrus F1populations generated by the following crosses: (A) ‘Pazit’ mandarin X ‘Chandler’ pummelo, and (B) ‘Ora’ mandarin X ‘Hudson’ grapefruit. Since our previous work demonstrated that furanocoumarin levels in the leaves is correlated to that in the fruit, we were able to characterize the F1 progeny at a very early stage for differences infuranocoumarin content, gene sequence and expression. Integration of the data lead to identification of candidate genes for key pathway enzymes, which were further analyzed for function using in-vivo transient expression.
Results
The ‘Pazit’ X ‘Chandler’ F1 population didn’t segregate for furanocoumarin levels (Figure 1)and therefore had a limited use for the identification of candidate genes for key enzymes. However, the ‘Ora’ X ‘Hudson’ population displayeda striking 50:50 segregation – half of the progeny produced significant levels of furanocoumarins, whereas the other half didn’t produce these compounds at all, or showed trace levels. This 50:50 segregation indicated that furanocoumarins biosynthesis is probably controlled by a single gene (Mendelian inheritance), of which ‘Ora’ contributed a ‘defective’ allele to each of the progeny, while ‘Hudson’ contributeda ‘defective’ allele to half of the progeny and a functional allele to the other half (Figure 1).
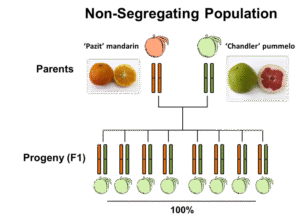
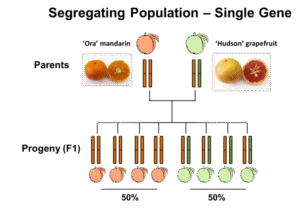

Figure 1. Two citrus populations discussed in this study
In order to identify the key gene involved in furanocoumarin biosynthesis in citrus fruit/leaves, we examined gene expression in ‘Ora’ vs. ‘Hudson’ young leaves.Based on studies conducted in other plants, we speculated that this gene likely belongs to the 2OGD family, which consists of 170 related genes in citrus. Oneprominent 2OGD gene was found to be highly expressed in ‘Hudson’ yet was absentfrom the’Ora’ transcriptome (‘Gene 710’). Examination of the gene structure in mandarin vs.pummelogenomes (no available grapefruit genome to date) revealed a 655 base insertion disrupting the mandarin gene, whereas the pummelo gene was found to be intact (Figure 2). Further analyses of other citrus varieties had shown an intact gene in sour orange, citron and lemon, while the same 655 base disrupting insertion was found in sweet orange. Next, an examination of the ‘Ora’ X ‘Hudson’ progeny revealed a full correlation between an intact allele presence and furanocoumarin levels. For extra validation, we tested the ‘Pazit’ X ‘Chandler’ progeny as well, and they all had an intact allele, in accordance with their significant furanocoumarin levels.
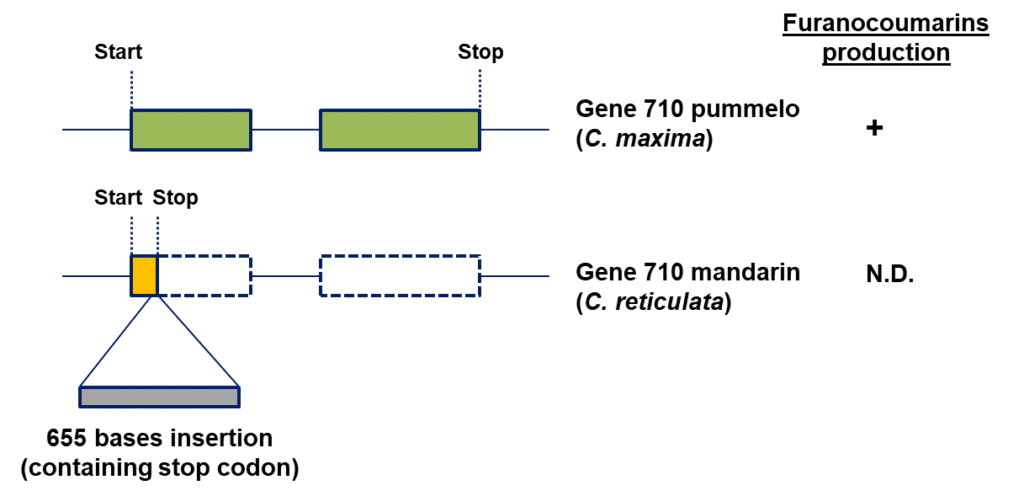
Figure 2.Gene 710 is intact in pummelo yet disrupted by a 655 base insertion in mandarin leading to an inactivated gene.
To confirm that ‘Gene 710’ is actually involved in furanocoumarins biosynthesis, we transiently expressed ‘Gene 710’ from pummelo in tobacco leaves and examined the activity of the encoded enzyme after adding suitable substrates. This experiment resulted with two products, scopoletin and umbelliferone (Figure 3), the latter being the first compound of the furanocoumarin biosynthesis pathway. We concluded that ‘gene 710’encodes a 2OGDinvolved in the first committed step in citrus furanocoumarin biosynthesis.
B.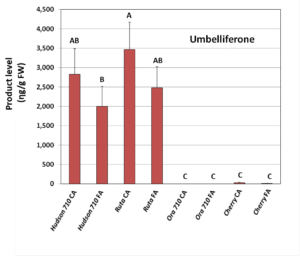
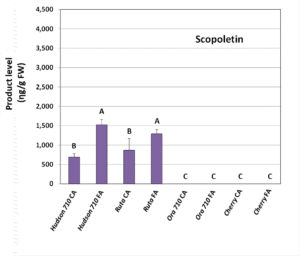
Figure 3. Activity assay product levels (umbelliferon-A and scopoletin-B) after expression of Hudson’s ‘Gene 710’, positive control from a different plant (Rutagraveolens), Ora’s ‘Gene 710’ and negative control (Cherry fluorescent protein) in tobacco leaves. Substrates were coumaric acid (CA) or ferulic acid (FA). Shown are means ± SE of three replicates. Different letters indicate statistically significant differences in product content (Fisher’s LSD test, P < 0.05).
A more detailed analysis of the genomic region around ‘Gene710’ hasrevealed that this gene occurs asone of five genes constituting a 2OGD gene cluster(Figure 4). Three of these genes are almost identical (Genes ‘700’, ‘710’ and ‘720’) whereas the other two flanking genes are less similar to ‘Gene 710’. Amongst the three almost identical genes, only ‘Gene710’ is expressed highly in ‘Hudson’ young leaves and fruitlets, and therefore is the main gene involved infuranocoumarin biosynthesis in citrus fruit/leaves.
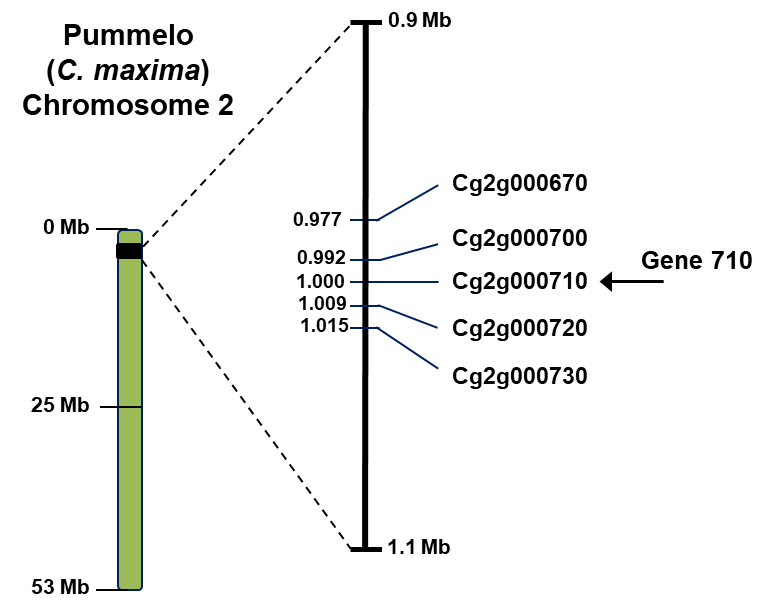
Figure 4. 2OGD gene cluster on pummelo chromosome #2. Genes 700 and 720 are almost identical to ‘Gene 710’ (~95%), whilegenes 670 and 730 are less similar (~85%).
We further studied the expression of the 2OGD gene cluster in other citrus tissues. Gene expression profile of both ‘Hudson’ and ‘Murcott’ (mandarin) roots showed that gene ‘720’ is the major gene involved in furanocoumarin biosynthesis in citrus roots (Figure 5).’Gene 730′ lacked elements of the substrate binding site and its expression was negligible; we therefore concluded that it is not functional., ‘Gene 670’ appeared to be expressed following pathogen infection and its activity was found to yieldonly one product, scopoletin but not umbelliferone (Figure 6), confirming that mandarin or orange fruit are not able to produce furanocoumarins, even after pathogen infection.
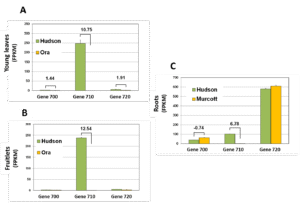
Figure 5. Expression of the three almost identical genes 700, 710 and 720 in young leaves (A) fruitlets (B) and roots (C) of grapefruit and mandarin.Shown are means ±SE of three replicates. DESEQ2 test in bold, Padj< 0.05.
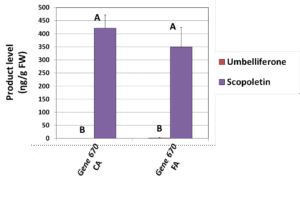
Figure 6. Activity assay products levels (umbelliferon and scopoletin) after expression of ‘Gene 670’in tobacco leaves. Substrates were coumaric acid (CA) or ferulic acid (FA). Shown are means ± SE of three replicates. Different letters indicate statistically significant differences in product content (Fisher’s LSD test, P < 0.05).
Conclusion
In the current study, we identified a gene that encodes a 2OGD that serves as a rate-limiting enzyme in furanocoumarin biosynthesis in citrus fruit. We demonstrated that it is functional in pummelo and grapefruit, but disrupted in mandarin and sweet orange. This gene is part of a 2OGD gene cluster, which includes a second gene involved in root-specific furanocoumarin biosynthesis and a third gene induced following plant-pathogen interactions.
Finally, our work provides the basis for future marker-assisted or genome-editing-based breeding of citrus varieties, which will be safe for consumption by a large and growing population of humans dependent on the use of prescription drugs.
Goldenberg, L., Ghuge, S.A., Doron-Faigenboim, A., Carmeli-Weissberg, M., Shaya, F., Rozen, A., Dahan, Y., Plesser, E., Kelly, G., Yaniv, Y., Arad, T., Ophir, R., Sherman, A., Carmi, N. and Eyal, Y. (2025), A 2OGD multi-gene cluster encompasses functional and tissue specificity that direct furanocoumarin and pyranocoumarin biosynthesis in citrus. New Phytol, 245: 1547-1562. https://doi.org/10.1111/nph.20322
igure 1. Two citrus populations discussed in this study
Photo 1 : Mandarin tree, var. ‘Shani’ (Photographs taken by Yossi Yaniv, ex.citrus orchard manager at ARO, Beit Dagan).




